Molecules that can hop
Chemically-fuelled mobile molecules, which perform mechanical work, underpin life in all its forms. An example is kinesin, a small protein that walks on microtubule filaments to transport cellular cargos or organize chromosomes during cell division. Researchers have been trying for decades to artificially reproduce these systems, aiming to attain their most notable feature: processivity.
Processivity is the ability to produce a useful action while moving directionally and continuously along a track. In an example from Chemical Biology, DNA polymerases synthesise DNA (useful work) while sliding along a template-chain (the track) dissociating from it only infrequently.
Despite the considerable challenge, Dr. Yujia Qing, and her colleagues Dr. Sandra A. Ionescu and Dr. Gökçe Su Pulcu from the group of Professor Hagan Bayley here at Oxford, succeeded in developing a one-legged mobile molecule: a molecular hopper (Fig. 1). The hopper is able to move directionally under external control, with pre-defined start and end points without the need for a supplemental chemical fuel. In addition, the hopper can carry a cargo (here a DNA) and even more amazingly, the system presents robust processivity.
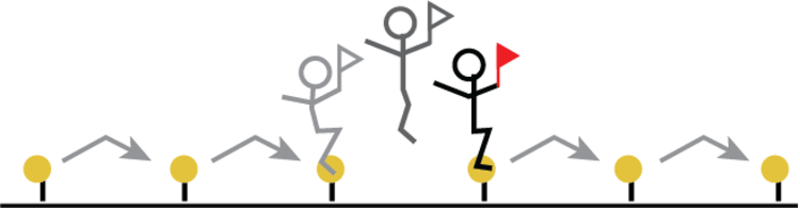
Fig.1 Schematic representation of the hopper’s movement.
The track along which the hopper moves is built into a protein nanopore, α-hemolysin (αHL), with cysteine footholds evenly distributed along the nanopore interior (Fig. 2A).
The movement of the hopper is controlled by the electric field inside the protein nanopore. The DNA cargo carried by the hopper has a negatively charged phosphodiester backbone, which experiences a pulling force under an applied potential. The force sets the direction of movement by aligning the hopper, which is track-bound through a disulfide bond, with the next cysteine foothold. (Fig .2B).
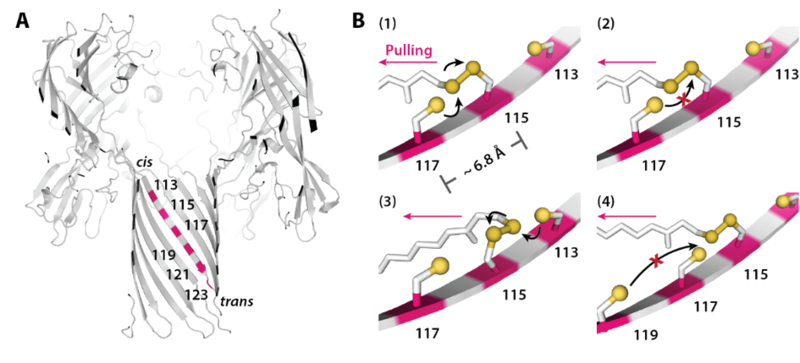
Fig. 2 (A) A six-foothold track comprising cysteine residues on a β strand of the αHL protein nanopore. (B) When an electrical potential is applied, the force on the DNA cargo aligns the disulfide bond connecting the hopper to the track with the downstream cysteine foothold, which initiates an interchange reaction (1). The formation of a disulfide bond between adjacent footholds on the track was not possible (2); backstepping was possible but rarely observed (3); no overstepping was observed (4).
Interestingly, one can induce the hopper to switch direction on the track by reversing the applied potential. For example, under a positive potential, the hopper moves downwards (in Fig. 4); switching to a negative potential re-orients the DNA and initiates upward movement. Although the cysteine footholds are close to each other on the track, no hopper release through disulfide bond cleavage and no overstepping were observed (Fig 2).
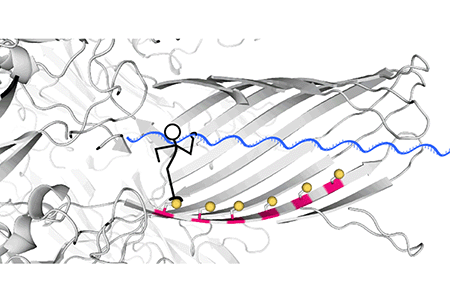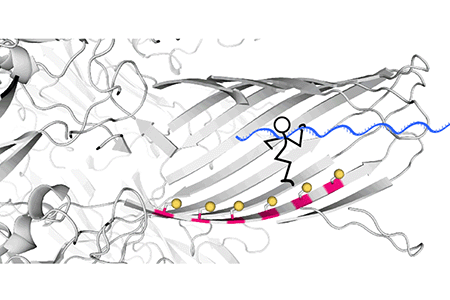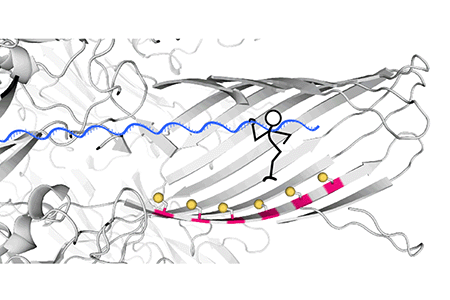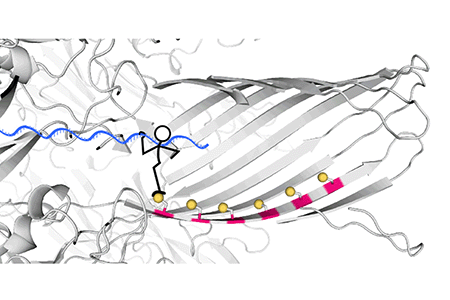
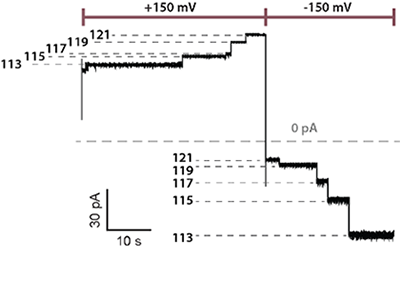
Fig. 3. Real-time recording of the hopper movement on a five-foothold track with a 40-nucleotide DNA cargo. As the hopper moved stepwise from one end of the track to the other, in either direction, each forward step made by the hopper increased the ion flow through the pore, producing staircase patterns of current.
At this point several questions arise, for example: How can the starting point be set up? How can the hopper’s motion be followed?
First, the hopper is delivered to the track by a carrier complex (Fig. 4). When the carrier-hopper enters the nanopore, the disulfide linkage between the hopper and the carrier becomes spatially with foothold Cys-115, and the ensuing reaction loads the hopper at a specific starting point. The traptavidin bound to the end of the carrier has a fundamental role in preventing the DNA from going through the nanopore without reacting.
To monitor the position of a hopper in real-time, Dr. Qing recorded the ionic current passing through a single protein nanopore under an applied potential. The ion flow through the nanopore was modulated to an extent dependent on the position of the hopper. As the hopper approached either end of the track, less DNA remained in the pore and a larger current was detected (Fig. 3).
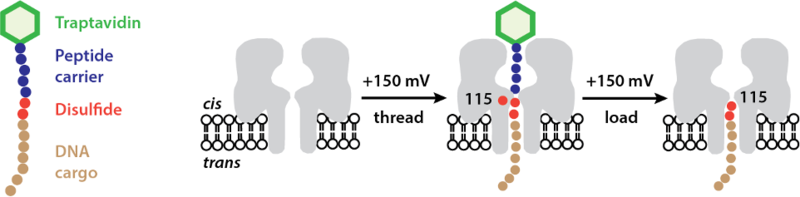
Fig. 4 The hopper is delivered to the track by a peptide carrier bound to a bulky protein, traptavidin, which prevents rapid translocation of the hopper through the nanopore and ensures the efficient loading of the hopper by a chemical reaction.
Starting from a DNA or RNA strand, single-molecule sequencing allows reading of nucleobases one by one in a sequence and thus identification of base modifications as well. The hopper system suggests a novel chemical ratcheting mechanism for single-molecule biopolymer sequencing. Its simplicity in design can potentially be transferred to a 2D surface for massively parallel ratcheting of any kind of polymer of interest. The immediate substrates are polypeptides and polysaccharides, sequencing of which will open up vast possibilities. Moreover, it demonstrates how Chemistry together with Biology and Bioengineering can help develop and upgrade technologies useful for medical investigation.
Article by Giorgia Fiorini
Image credit (hopping bird): Maomao Zhang