Hagan Bayley

Professor Hagan Bayley FRS
Professor of Chemical Biology
Our laboratory uses the techniques of protein chemistry, molecular genetics, biophysics and cell biology. Much of our work is centered on membrane proteins, in particular channels and pores. We investigate both the fundamental properties of these proteins and their applications in biotechnology. Several of our studies are collaborations with other laboratories in the UK, USA and Europe. There are currently 10 postdocs, 12 graduate students and a technician in the laboratory. They are from 12 different countries and 15 are women.
Printed synthetic, cellular and hybrid tissues

We discovered a process by which aqueous droplets can be connected by means of lipid bilayers to form networks. Protein pores incorporated into the bilayers allow the droplets to communicate with each other and the environment. In the area of synthetic biology, considerableeffort has been devoted to the preparation of artificial cells. By contrast, assemblies of interacting compartments, which act as tissues, have been hardly explored. Droplet networks are an advance in this direction. By using engineered pores in the interface bilayers, we have produced droplet networks that form batteries, detect light and rectify electrical signals. Recently, we have built more extensive networks by 3D printing to form synthetic tissues that conduct signals along neuron-like pathways or fold to assume altered shapes. The ability to subject synthetic tissues to external control, e.g. with light or magnetism, is an active interest. Further, the printing technology has been adapted to pattern living cells in three-dimensional patterns, enabling the fabrication of hybrid synthetic/ cellular materials.
Research papers:
Villar, G., Graham, A.D. and Bayley, H. A tissue-like printed material. Science 340, 48-52 (2013). DOI: 10.1126/science.1229495
Booth, M.J. Restrepo Schild, V., Graham, A.D., Olof, S.N. and Bayley, H. Light-activated communication in synthetic tissues. Science Advances 2, e1600056 (2016). doi:10.1126/sciadv.1600056
Review:
Booth, M.J., Restrepo-Schild, V., Downs, F. and Bayley, H. Functional aqueous droplet networks. Mol. BioSystems 13, 1658- 1691 (2017). DOI: 10.1039/c7mb00192d
Polypeptide translocation and sequencing
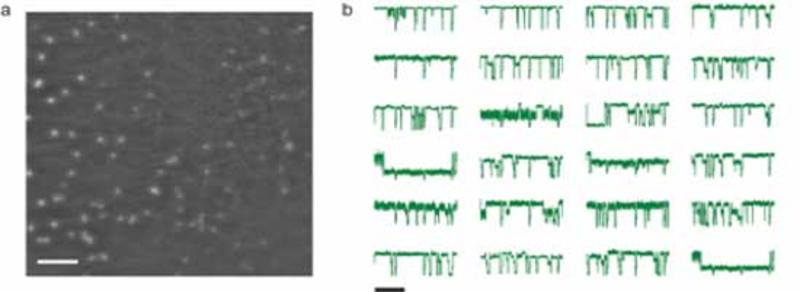
We have engineered many variants of the α-hemolysin protein pore for single molecule detection (stochastic sensing), which is effected by monitoring the modulation of an ionic current passing through a single pore. Stochastic sensing allows the analysis of a wide variety of analytes: metal cations, small organic molecules, nucleic acids, proteins, &c. The spin-out company from the Bayley laboratory, Oxford Nanopore, has used this technology to sequence single molecules of DNA and RNA (https://www.nanoporetech.com/). Following the success of Oxford Nanopore's MinION sequencer, we have redirected our efforts towards protein characterization, in particular the detection of post-translational modifications and alternative splicing. To pass through a nanopore, a protein must unfold so that the polypeptide chain can thread through the narrow lumen. Accordingly, this process is also revealing fundamental information about protein folding and the trafficking of proteins between compartments in cells. A critical issue in stochastic sensing is the implementation of highly parallel analyte detection, and we are currently developing technologies to allow simultaneous recording from thousands of protein nanopores.
Research papers:
Rosen, C.B., Rodriguez-Larrea, D. and Bayley, H. Single-molecule site-specific detection of protein phosphorylation. Nature Biotechnology 32, 179-181 (2014). doi:10.1038/nbt.2799
Huang, S., Romero-Ruiz, M., Castell, O.K., Bayley, H., and Wallace, M.I. High-throughput optical sensing of nucleic acids in a nanopore array. Nature Nanotechnology 10, 986-991 (2015). DOI: 10.1038/NNANO.2015.189
Review:
Bayley, H. Nanopore sequencing: from imagination to reality. Clinical Chemistry 61, 25-31 (2015). DOI: 10.1373/clinchem.2014.223016
Single molecule chemistry and catalysis

Protein pores can be used as nanoreactors to study covalent chemistry at the single-molecule level. We have investigated a wide variety of chemistry in this way, including the formation and cleavage of arsenic-sulfur bonds, the photochemistry of 2-nitrobenzyl protecting groups, the photoisomerization of azobenzenes, the observation of polymerization one step at a time and a hydrogen-deuterium isotope effect. New directions include the incorporation of unnatural amino acids into the a-hemolysin pore, with which to expand the range of chemistries that can be investigated, and the examination of catalysis. The nanoreactor technology is also being adapted to study complex reaction networks and molecular walkers. Recently, we have demonstrated a processive molecular hopper that steps with nanometer resolution and reverses on command.
Research papers:
Pulcu, G.S., Mikhailova, E., Choi, L.-S. and Bayley, H. Continuous observation of the stochastic motion of an individual small-molecule walker. Nature Nanotechnology 10, 76-83 (2015). DOI: 10.1038/NNANO.2014.264
Qing, Y., Ionescu, S.A., Pulcu, G.S. and Bayley, H. Directional control of a processive molecular hopper. Science, in press (2018).
Review:
Bayley, H., Luchian, T., Shin, S.-H. and Steffensen, M.B. Single-molecule covalent chemistry in a protein nanoreactor, pp251-277, Chapter 10 in "Single Molecules and Nanotechnology", R. Rigler and H. Vogel, eds., Springer, Heidelberg (2008)
Engineered nanopores
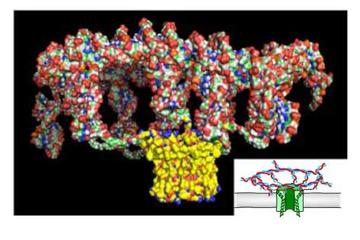
We continue to engineer new pores for applications in biotechnology, including sensing and sequencing, single-molecule chemistry and the fabrication of synthetic tissues. Recent examples include pores containing unnatural amino acids assembled by native chemical ligation, pores assembled from synthetic peptides, and truncated pores that nonetheless form openings in lipid bilayers. In a new venture, we have prepared DNA nanostructure-polypeptide hybrids that form large pores of defined diameter.
Research papers:
Stoddart, D., Ayub, M., Höfler, L., Raychaudhuri, P., Klingelhoefer, J.W., Maglia, G., Heron, A. and Bayley, H. Functional truncated membrane pores. Proc. Natl. Acad. Sci. USA 111,2425-2430 (2014) www.pnas.org/cgi/doi/10.1073/pnas.1312976111
Spruijt, E., Tusk, S.E. and Bayley, H. DNA scaffolds support stable and uniform peptide nanopores. Nature Nanotechnology 13, 739-785 (2018). doi.org/10.1038/s41565-018-0139-6
Review:
Ayub, M. and Bayley, H. Engineered transmembrane pores. Curr. Op. Chem. Biol. 34, 117-126 (2016). http://dx.doi.org/10.1016/j.cbpa.2016.08.005
Hagan Bayley is the Professor of Chemical Biology at the University of Oxford. A major interest of his laboratory is the development of engineered protein nanopores for stochastic detection, including single-molecule covalent chemistry and ultrarapid biopolymer sequencing. Recently, the Bayley lab has developed techniques for the fabrication of 3D tissues, both living and synthetic. In 2005, Professor Bayley founded Oxford Nanopore, which has manufactured the portable MinION and other DNA/RNA sequencers.
Professor Bayley was the 2009 Chemistry World Entrepreneur of the Year. In 2011, he was elected a Fellow of the Royal Society. In 2012 he was awarded the Royal Society of Chemistry's Interdisciplinary Prize, in 2017 the Menelaus Medal of the Learned Society of Wales and in 2019 the Mullard Award of the Royal Society. In 2018, Professor Bayley held the Kavli Chair at the Delft University of Technology and he is currently a Program Director of the Oxford Martin School.
Contact
01865 285100




