Light-driven protein modification
Post-translational modifications (PTMs) are chemical modifications occurring inside the cell that increase the structural and functional diversity of the proteome; for example, by modification of an existing functional group on the protein. These modifications play important roles in numerous cellular processes and can be used for the analysis of natural systems and the development of biological tools. Despite the advantages that protein modification can offer, PTMs and the synthetic strategies that mimic them can still be limited in the range of modifications that can be carried out and the efficiency of such processes.
Researchers from the groups of Prof. Veronique Gouverneur and Prof. Benjamin Davis have developed an alternative strategy to address these limitations involving the post-translational functionalization of proteins. Using visible light, carbon-centered radicals, which are highly reactive chemical species, can be generated to carry out site-selective protein editing through the formation of carbon–carbon bonds with minimal protein damage (Figure 1). These reactions proceed under mild conditions and with high efficiencies, providing a diverse range of proteins with functionalities that would otherwise be difficult to access.
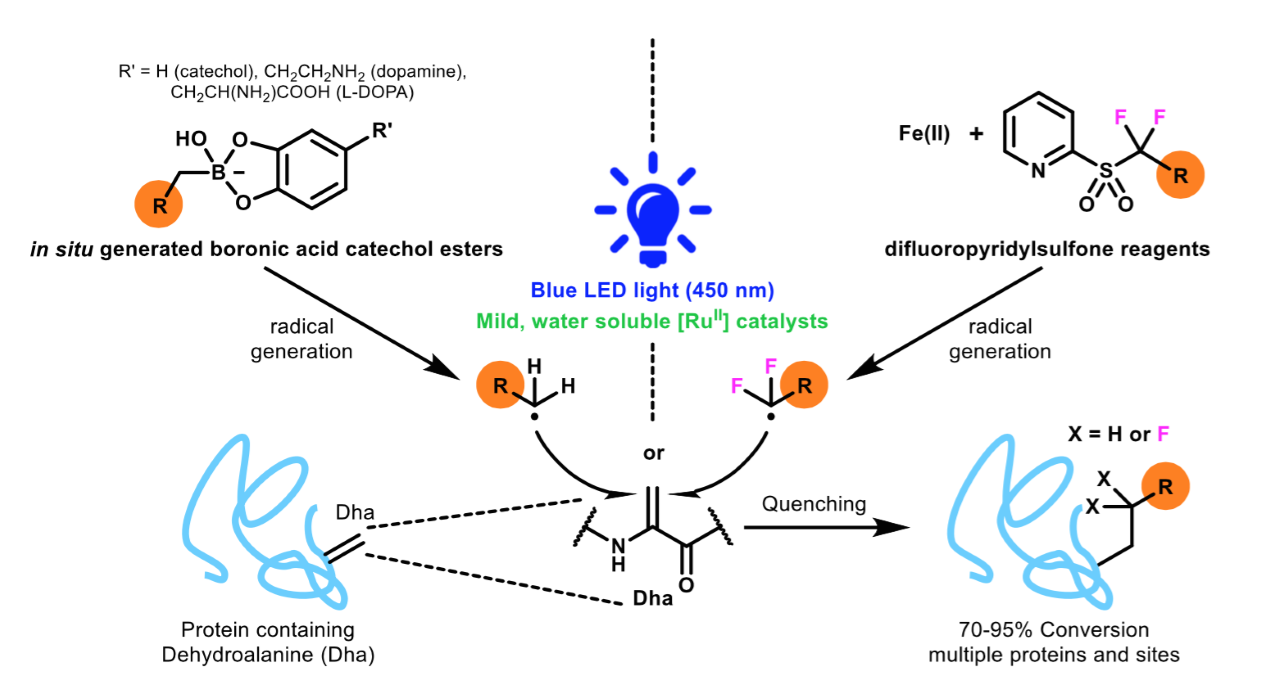
Figure 1: Generating carbon-centered radicals with light for protein modification.
This work is the culmination of an extensive interdisciplinary collaboration that began with an investigation into light-based synthetic strategies for radical reactions in water. Although visible light has been used extensively to promote radical reactions with small molecules in non-aqueous environments, its applications for the modification of large biomolecules has been more limited.
Thorough optimization and control of the reaction conditions, the electron transfer processes to form the reactive radical species and the design of the radical precursors resulted in the development of a protocol to form radicals that can react efficiently with dehydroalanine residues on proteins. These residues were targeted as they are readily generated and are highly reactive towards radical species by acting as a radical acceptor.
A bespoke class of chemical reagents was also developed as a precursor to fluorine-containing radicals to enable the installation of fluorinated motifs. The selective incorporation of fluorine labels onto proteins is highly desirable, as it would enable the use of techniques such as fluorine nuclear magnetic resonance (NMR) spectroscopy to probe protein structure and dynamics.
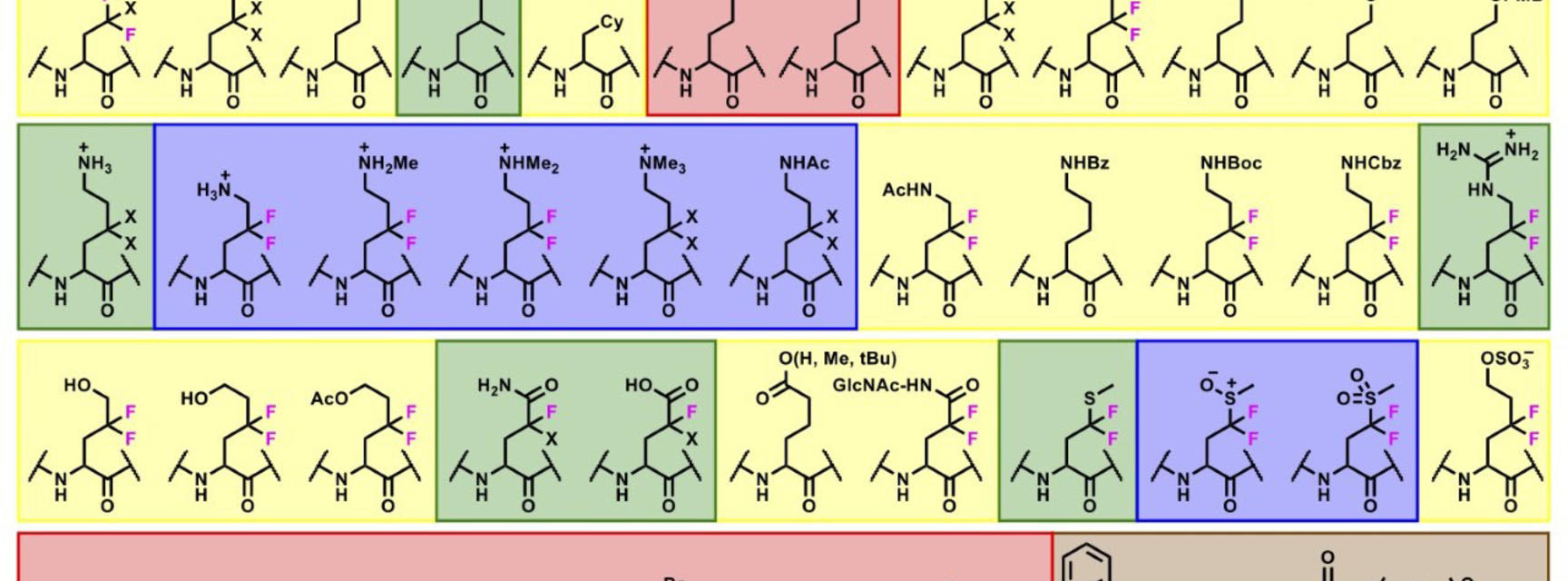
Over 50 unique side chains could be installed onto a diverse range of protein scaffolds and sites with this synthetic methodology, displaying an impressive chemical tolerance (Figure 2). The reaction can also be initiated in the presence of reactive groups on the radical precursors, broadening the scope of functional groups that can be appended onto the protein scaffold. By installing side chains that themselves contain a radical precursor, on-protein radicals can be formed under certain conditions and can undergo further reactions to access more complex structures (Figure 3).
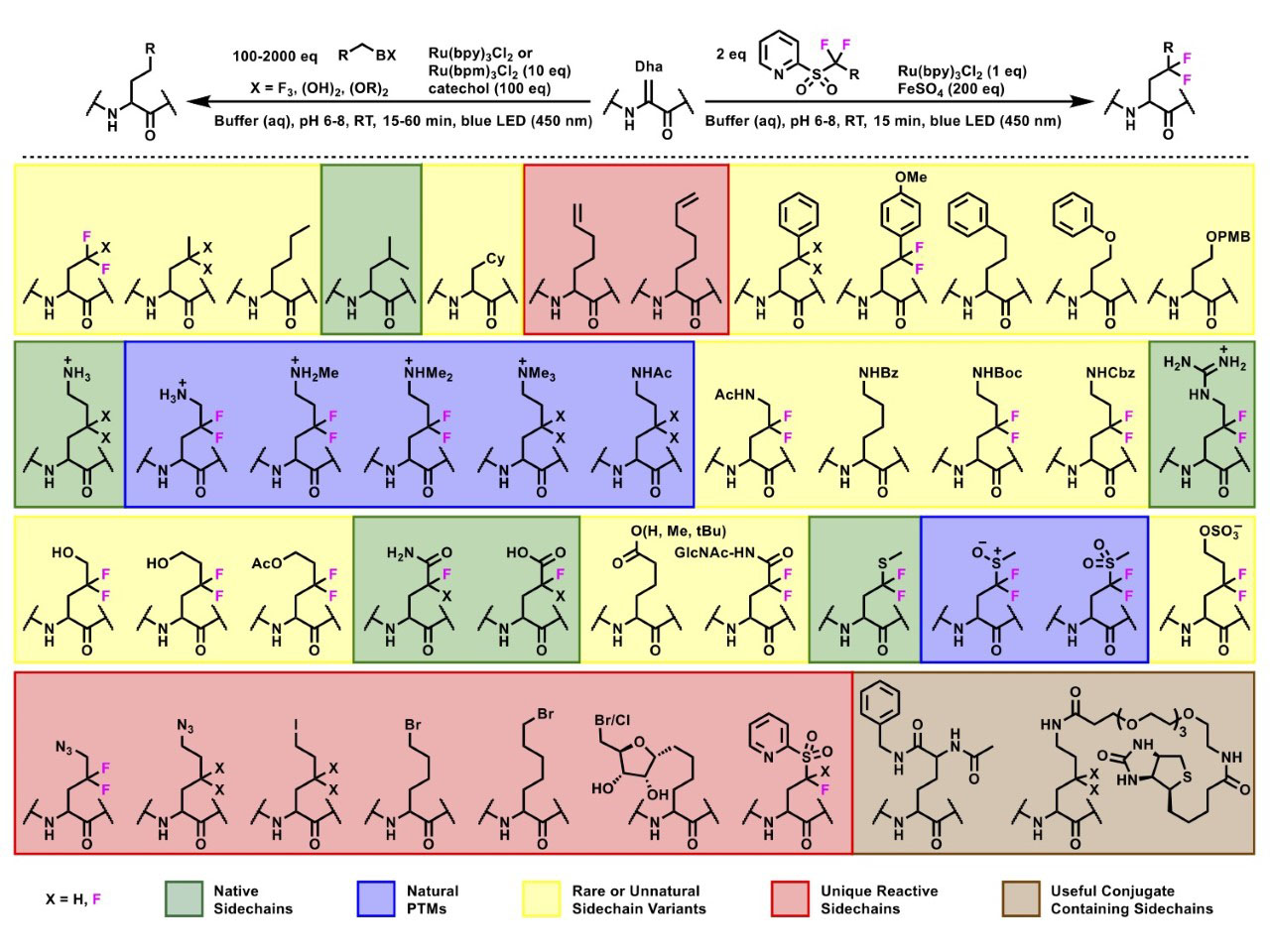
Figure 2: Substrate scope of light-driven protein modification
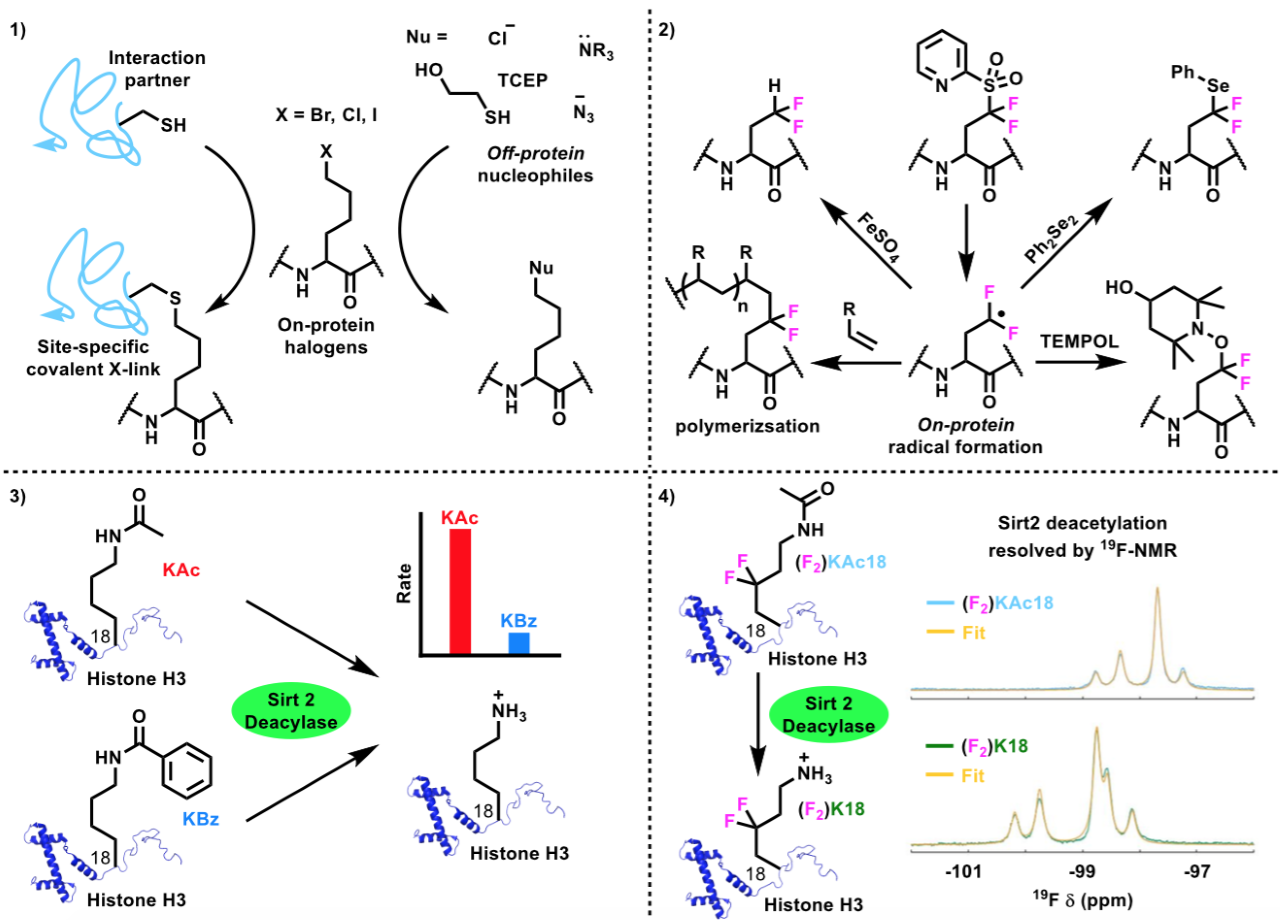
Figure 3: Novel applications of previously inaccessible side-chains.
This methodology successfully overcomes the many challenges associated with developing efficient tools for biomolecule modification, including the requirement for mild reaction conditions that are non-destructive to biological substrates, compatibility with water as a reaction medium and the ability to achieve high selectivity for the reaction in the presence of a variety of other functional groups that can be found in a biological environment.
As one of the most efficient ways available to selectively alter the structure of proteins, this work opens up vast possibilities for more complex and interesting chemical modifications of a variety of biological systems. The incorporation of reactive motifs that are not found in nature imparts completely novel reactivity in biomolecules and has the potential to guide further discoveries that will push the boundaries of protein chemistry.
Article by Marie Wong. Many thanks to Brian Josephson and Dr. Patrick Isenegger for help with this article.
Image credit: Brian Josephson.