A novel NMR technique developed in Oxford to study the interactions between cells and pathogens such as influenza and SARS-CoV-2 has been published today in Science.
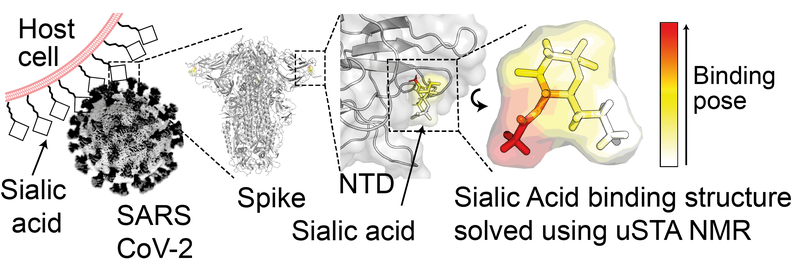
Figure 1: Sugar-pathogen binding confirmed with the novel method of uSTA NMR.
Using this method a team led by the Baldwin, Davis, and Naismith groups were able to confirm a novel binding mode between the spike protein on the original strain of SARS-CoV-2 and specific sialoside (sugar) residues on proteins inside lung cells. As SARS-CoV-2 mutated, the more infective variants (alpha, beta, delta, omicron) lost this binding function. The team, made up of researchers from Oxford Chemistry and the Kavli Institute for Nanoscience Discovery, as well as collaborators in the Wellcome Centre for Human Genetics and the Rosalind Franklin Institute, were able to track this evolution using their NMR method.
When a virus enters a cell a complex series of recognition events occur, often relying on interactions between the viral proteins and particular glycans (sugars) on the surface of the host cell (see Fig. 1). Analysing these interactions, either biochemically or using NMR, is important for tracking the spread and evolution of these viruses. For many protein systems however, including SARS-CoV-2, the interactions are difficult to analyse because of the complex protein structures and wide variety of possible interactions.
In a study published today, the team devised an NMR method that can robustly, rapidly, and quantitatively analyse these interactions for a range of systems, including those previously inaccessible to existing NMR methods.
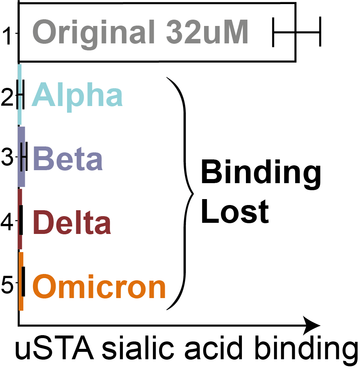
Figure 2: Binding lost in more transmissible variants.
Building on previous protein-NMR techniques, the team’s new method of “universal saturation transfer analysis” (uSTA) uses a thorough mathematical model of the magnetization transfer between the virus and host, providing a wealth of information on exactly how viral proteins interact with cell-surface sugars. Their automated and openly available workflow can be readily applied in a range of systems, and allows researchers to obtain quantitative binding parameters for previously intractable systems.
Alongside this work, genetic analysis from patients early in the Covid-19 pandemic uncovered variations in their cell glycolysation that may be relevant to the binding mode the team identified, potentially contributing to variations in disease severity.
Professor Ben Davis, one of the paper’s senior authors, said:
Two of the ongoing mysteries of the coronavirus pandemic are the mechanisms behind viral transmission and the origins of the zoonotic leap.
There is evidence that some influenza viruses can grab sialic acid on the surface of human host cells, and this has been seen in Middle Eastern Respiratory Syndrome (MERS), which is a coronavirus. Although SARS-CoV-2 variants of concern had not shown this mechanism, our research finds that the viral strain that emerged in early 2020 could use this as a way of getting into human cells.
You can read more about the study in Science here.